DOI: 10.1039/D2EN01077A (Paper) Environ. Sci.: Nano, 2023, 10, 2245-2258
Abstract
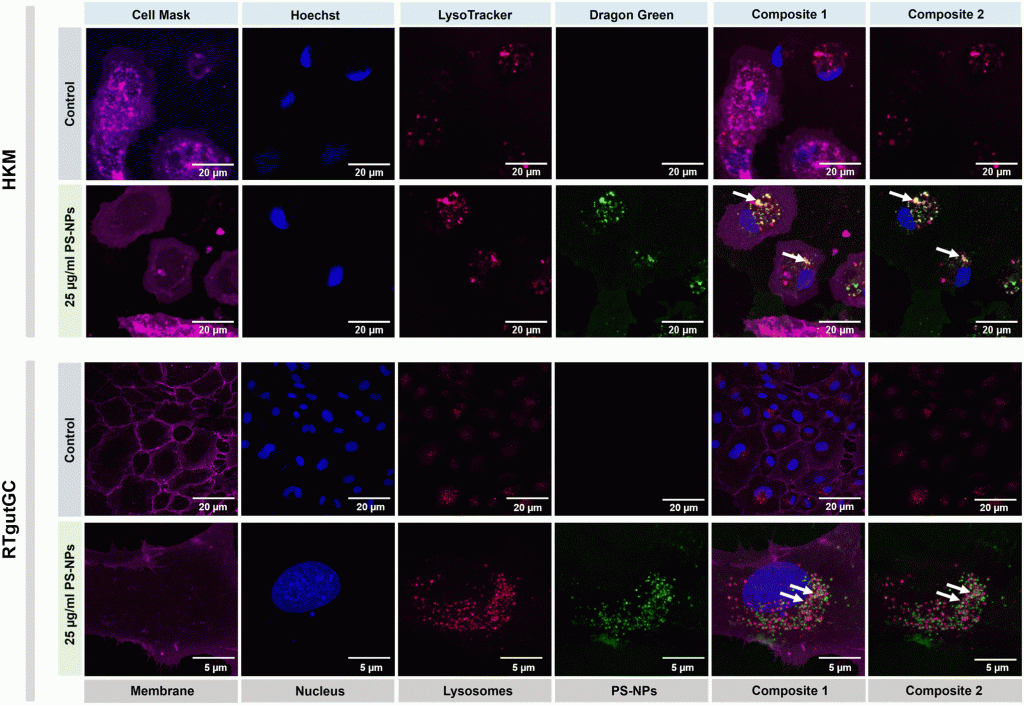
Nanoplastics (NPs) are currently a main concern for environmental, animal and human health due to their potential to accumulate in different environmental compartments and provoke effects in living organisms. Nevertheless, neither these effects nor the interaction of NPs with the cellular machinery are well characterized, and only scattered information is available. In the present work, we focused on the interaction between NPs and fish cells, both intestinal cells and macrophages, in order to understand which cell organelles are targeted by polystyrene (PS)-NPs and how this could impact cell function. PS-NPs can pass through phospholipid membranes, entering cells via endocytosis, phagocytosis or passive transport. Once internalized, we found that PS-NPs co-localize with lysosomes but not with mitochondria. Moreover, using two types of fluorescent probe (H2DCFDA and DHE) we demonstrated that NPs did not trigger the production of reactive oxygen species (ROS), which was corroborated by the fact that neither the oxidative consumption ratio (OCR) nor the extracellular acidification rate (ECAR) in mitochondrial respiration were altered. RNASeq data revealed clear interference by PS-NPs with lipid metabolism, peroxisomes and PPAR signaling. The M1/M2 balance critically determines tissue homeostasis when exposed to exogenous agents such as microorganisms or pollutants. Thus, the expression of different genes (il1β, tnfα, il6, il10, il12, cox2, mmp9, ppar a, b and g) was further assessed to characterize the macrophage phenotype M1 or M2, induced by PS-NPs. Overall, in this study we demonstrate that PS-NPs co-localize within lysosomes, both in macrophages and in intestinal cells of rainbow trout, but do not trigger ROS production nor alter mitochondrial respiration. In macrophages, PS-NPs modulate polarization towards the M2-like phenotype.