Our lab uses protein crystallography with synchrotron radiation as a major procedure to decipher the molecular mechanisms that lay behind the atomic structure of proteins and protein complexes. In our lab we combine this powerful structural technique with a functional and biochemical characterization using either in vitro or in vivo methods. In the last decades protein-function characterization of proteins and protein complexes have shed light into the most relevant discoveries in biochemistry and molecular biology.
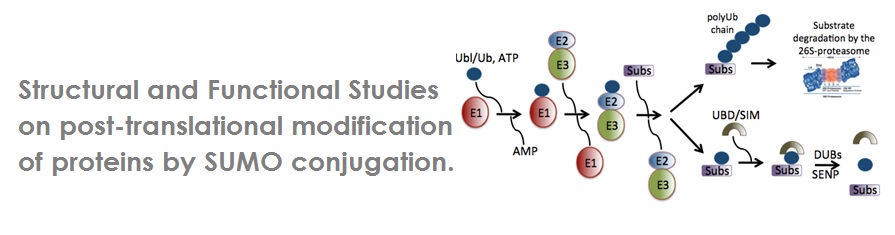 SUMO and ubiquitin are small protein modifiers that can be attached via an iso-peptidic bond to lysine residues of target proteins. This type of post-translational modification is very common and regulate almost all processes of cell life, including cell division, DNA repair or gene expression. For example, ubiquitin modification through Lys48 regulates the half-life of many proteins by degradation with the proteasome system and is essential for the protein homeostasis in the cell.
SUMO and ubiquitin are small protein modifiers that can be attached via an iso-peptidic bond to lysine residues of target proteins. This type of post-translational modification is very common and regulate almost all processes of cell life, including cell division, DNA repair or gene expression. For example, ubiquitin modification through Lys48 regulates the half-life of many proteins by degradation with the proteasome system and is essential for the protein homeostasis in the cell.
The conjugation of ubiquitin and SUMO (Ubl) to target proteins is conducted via a conserved multistep enzymatic cascade through E1 (activating enzyme), E2 (conjugating enzyme), and E3 (ligase enzyme). Reversely, deubiquitinating enzymes (DUBs) can remove ubiquitin by catalyzing the hydrolysis of the isopeptide bond. Therefore, ubiquitin and SUMO conjugation and deconjugation are balanced and tightly regulated by E3 ligases conjugation and DUBs deconjugation.
Structural/Functional Characterization of the USP25 deubiquitinating enzyme
Human USP25 (and USP28) are deubiuquitinating proteases that control the levels of important targets in the cell and are regulated, among other systems, by SUMO conjugaiton on the N-terminal domain. USP25 (and USP28) are modular proteins composed by three domains: a N-terminal regulatory domain that interact with the ubiquitin chains; a central USP-like domain with the catalytic residues and including a long insertion in the middle of the domain; and a C-terminal domain that interacts with specific substrates, such as the recently reported Tankyrases involved in the Wnt/β-catenin pathway.
We have recently solved the crystal structure of USP25, which reveals the presence of a homotetrameric structure which is involved in a novel regulatory mechanism of the deubiquitinating activity.
Liu, B., Sureda-Gómez, M., Zhen, Y., Amador, V., Reverter, D. (2018). A quaternary tetramer assembly inhibits the deubiquitinating activity of USP25. Nature Communications 9, 4973.
Structural/Functional Characterization of the SMC5/6 complex, a multimeric SUMO E3 ligase enzyme
SMC (Structural Maintenance of Chromosomes) complexes are topologically closed molecules formed by two elongated SMC subunits and by a distinct number of associated non-SMC elements (NSE). SMC proteins contain three different domains: an ATPase head structurally related to that of ABC transporters (hereafter named “HEAD”), an extended coiled coil region (“ARM”) and a heterodimerization or hinge domain (“HINGE”). Each SMC complex has specific and essential roles: cohesin maintains connections between sister chromatids, condensin compacts chromosomes and Smc5/6 promotes chromosome disjunction. Despite these seemingly disparate functions, all SMC complexes share a common property, which is to organize chromosomes by topologically embracing DNA inside their ring-shaped structure.
We have recently shown that the Nse2 SUMO E3 ligase in the Smc5/6 complex, a critical player during recombinational DNA repair, is directly stimulated by binding to DNA. Activation of SUMOylation requires the electrostatic interaction between DNA and a positively-charged patch in the ARM domain of the Smc5 subunit, which acts as a DNA sensor that subsequently promotes an stimulation of the Nse2 ligase activity. These results reveal a novel mechanism to enhance a SUMO E3 ligase activity by direct DNA-binding and to restrict SUMOylation in the vicinity of those Smc5/6-Nse2 molecules engaged on DNA.
Varejão, N., Ibars, E., Lascorz, J., Colomina, N., Torres-Rosell, J., Reverter, D. (2018). DNA activates the Nse2/Mms21 SUMO E3 ligase in the Smc5/6 complex. EMBO J. pii: e98306. (News and views, Pichler, A. (2018). How DNA vicinity controls SUMO E3 ligase activity. EMBO J. pii: e99705)
Temperature-dependent activation an hyperthermophilic esterase
Esterases and lipases are very important biocatalysts for industrial purposes, since they catalyze reactions of synthesis or hydrolysis of lipidic ester bonds. In general agreement, esterases (E.C. 3.1.1.1) prefer short to medium chains up to C10 of monoesters, whereas lipases (E.C. 3.1.1.3) can hydrolyze water-insoluble long-chain triglycerides. The Pf2001 esterase from Pyrococcus furiosus reaches its optimal activity between 70 and 80°C.
We have recently solved the crystal structure of the Pf2001 esterase, which shows two different conformations: monomer and dimer. The structures reveal important rearrangements in the “cap” subdomain between monomer and dimer, by the formation of an extensive intertwined helical interface. Moreover, the dimer interface is essential for the formation of the hydrophobic channel for substrate selectivity, as confirmed by mutagenesis and kinetic analysis. We propose a novel temperature-dependent activation mechanism of the Pf2001 esterase by dimerization.
Varejão, N., De-Andrade, R.A., Almeida, R.V., Anobom, C.D., Foguel, D., Reverter D. (2017). Structural Mechanism for the Temperature-Dependent Activation of the Hyperthermophilic Pf2001 Esterase. Structure. S0969-2126(17), 30403-30413.