 Researchers from the Bacterial Pathogenesis and Antimicrobials group of the Institut de Biotecnologia i de Biomedicina (IBB) and the Department of Genetics and Microbiology at UAB, in collaboration with the Research Center Borstel – Leibniz Lung Center (Germany) and ISGlobal at Hospital Clínic (Barcelona), have established a link between the quorum sensing system and the virulence and resistance phenotypes in Stenotrophomonas maltophilia, and identified potential high-risk clones circulating worldwide.
Researchers from the Bacterial Pathogenesis and Antimicrobials group of the Institut de Biotecnologia i de Biomedicina (IBB) and the Department of Genetics and Microbiology at UAB, in collaboration with the Research Center Borstel – Leibniz Lung Center (Germany) and ISGlobal at Hospital Clínic (Barcelona), have established a link between the quorum sensing system and the virulence and resistance phenotypes in Stenotrophomonas maltophilia, and identified potential high-risk clones circulating worldwide.
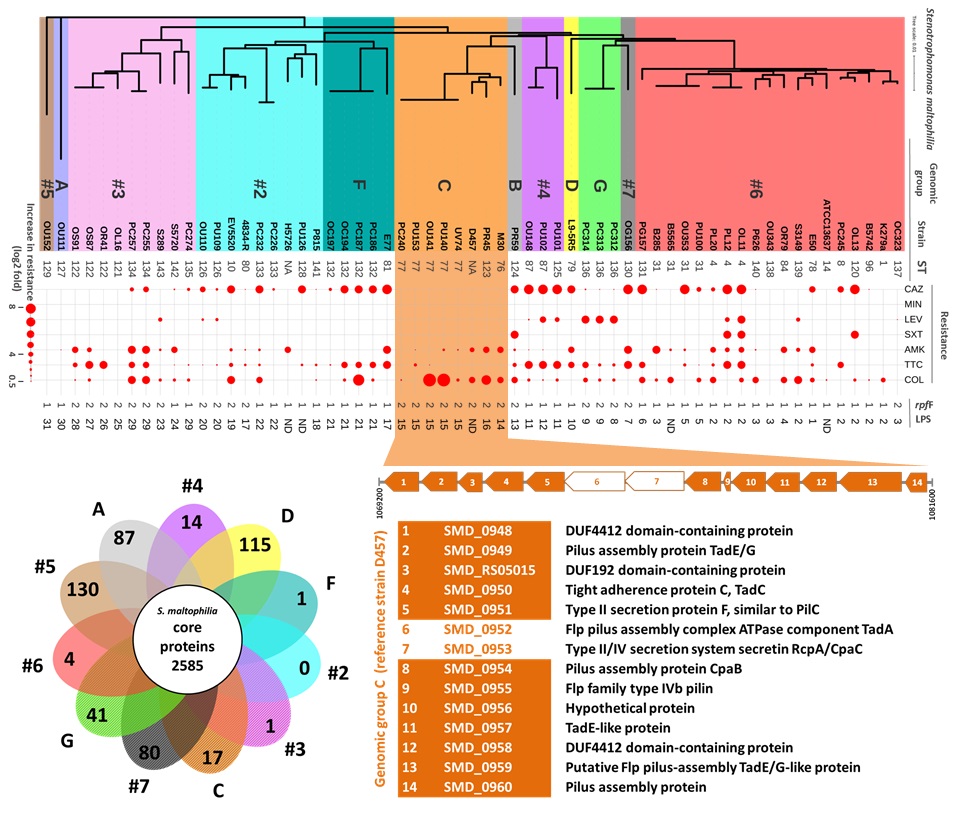
S.maltophilia are increasingly recognized as significant opportunistic pathogens in healthcare settings worldwide, the global spread of multidrug-resistant strains of this species being the most serious concern. Epidemiological studies are important to identify particular lineages or strains exhibiting clinically relevant phenotypes and to make knowledge-driven healthcare decisions.
In this context, the UAB group, in collaboration with research groups in Germany and Hospital Clínic, have studied the link between quorum sensing, virulence and antibiotic resistance in a panel of genetically diverse clinical S. maltophilia isolates from different European countries. In particular, a clonal group of strains were identified that show an increased ability to form biofilms and exhibit higher resistance to the last-resort antibiotic colistin. In addition, new virulence factors exclusive to this lineage have been identified through a comparative genomics study.
The results, published in Frontiers in Microbiology, demonstrate the pivotal role of the quorum sensing system in the pathogenicity and persistence in S. maltophilia and alert on the potential risk of resistant and virulent clones circulating in European hospitals.
In connection with this study, the UAB group has also participated in an ambitious project aimed to describe the population structure and spread of S. maltophilia at a global scale, led by the Research Center Borstel. The results of this project have been published in Nature Communications. The analysis of a worldwide collection of 1.305 isolates detected human-associated lineages with higher proportions of key virulence and resistance genes.
Link to paper in Frontiers in Microbiology:
https://www.frontiersin.org/articles/10.3389/fmicb.2020.01160/full
Link to paper in Nature Communications:
https://www.nature.com/articles/s41467-020-15123-0
