Nature Communications 10, Article number: 4222 (2019)
https://www.nature.com/articles/s41467-019-12173-x.epdf?author_access_token=cVjT1p3din4tGJbNLDyel9RgN0jAjWel9jnR3ZoTv0P-2IqhjSjJ3h8OoQACdg3_2Zs57EgxdseDbCAlUgZO9jSYZMidnZsRJxkAkaIfu2LAxppiY3VN8K2eAt2G6O7iHH_N5qqpM4UMjovqEWHu9A%3D%3D
Abstract
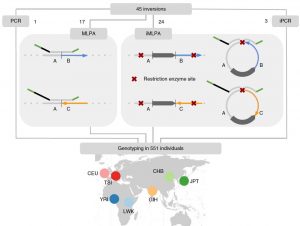 Inversions are one type of structural variants linked to phenotypic differences and adaptationin multiple organisms. However, there is still very little information about polymorphicinversions in the human genome due to the difficulty of their detection. Here, we develop anew high-throughput genotyping method based on probe hybridization and amplification, andwe perform a complete study of 45 common human inversions of 0.1–415 kb. Most inversionspromoted by homologous recombination occur recurrently in humans and great apes andthey are not tagged by SNPs. Furthermore, there is an enrichment of inversions showingsignatures of positive or balancing selection, diverse functional effects, such as gene dis-ruption and gene-expression changes, or association with phenotypic traits. Therefore, ourresults indicate that the genome is more dynamic than previously thought and that humaninversions have important functional and evolutionary consequences, making possible todetermine for thefirst time their contribution to complex traits
Inversions are one type of structural variants linked to phenotypic differences and adaptationin multiple organisms. However, there is still very little information about polymorphicinversions in the human genome due to the difficulty of their detection. Here, we develop anew high-throughput genotyping method based on probe hybridization and amplification, andwe perform a complete study of 45 common human inversions of 0.1–415 kb. Most inversionspromoted by homologous recombination occur recurrently in humans and great apes andthey are not tagged by SNPs. Furthermore, there is an enrichment of inversions showingsignatures of positive or balancing selection, diverse functional effects, such as gene dis-ruption and gene-expression changes, or association with phenotypic traits. Therefore, ourresults indicate that the genome is more dynamic than previously thought and that humaninversions have important functional and evolutionary consequences, making possible todetermine for thefirst time their contribution to complex traits
