ARTICLE| VOLUME 28, ISSUE 2, P352-367.E9, JULY 09, 2019
Covadonga Vara; Andreu Paytuví-Gallart ; Yasmina Cuartero ;Paul D. Waters; Marc A. Marti-Renom; Aurora Ruiz-Herrera
Open Access DOI: https://doi.org/10.1016/j.celrep.2019.06.037
https://www.cell.com/cell-reports/fulltext/S2211-1247(19)30803-4
 Summary
Summary
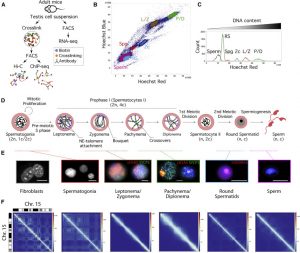
Mammalian gametogenesis involves dramatic and tightly regulated chromatin remodeling, whose regulatory pathways remain largely unexplored. Here, we generate a comprehensive high-resolution structural and functional atlas of mouse spermatogenesis by combining in situ chromosome conformation capture sequencing (Hi-C), RNA sequencing (RNA-seq), and chromatin immunoprecipitation sequencing (ChIP-seq) of CCCTC-binding factor (CTCF) and meiotic cohesins, coupled with confocal
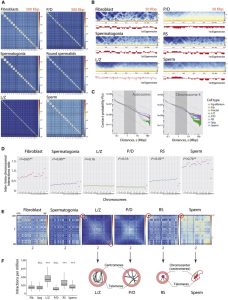
and super-resolution microscopy. Spermatogonia presents well-defined compartment patterns and topological domains. However, chromosome occupancy and compartmentalization are highly re-arranged during prophase I, with cohesins bound to active promoters in DNA loops out of the chromosomal axes. Compartment patterns re-emerge in round spermatids, where cohesin occupancy correlates with transcriptional activity of key developmental genes. The compact sperm genome contains compartments with actively transcribed genes but no fine-scale topological domains, concomitant with the presence of protamines. Overall, we demonstrate how genome-wide cohesin occupancy and transcriptional activity is associated with three-dimensional (3D) remodeling during spermatogenesis, ultimately reprogramming the genome for the next generation.
